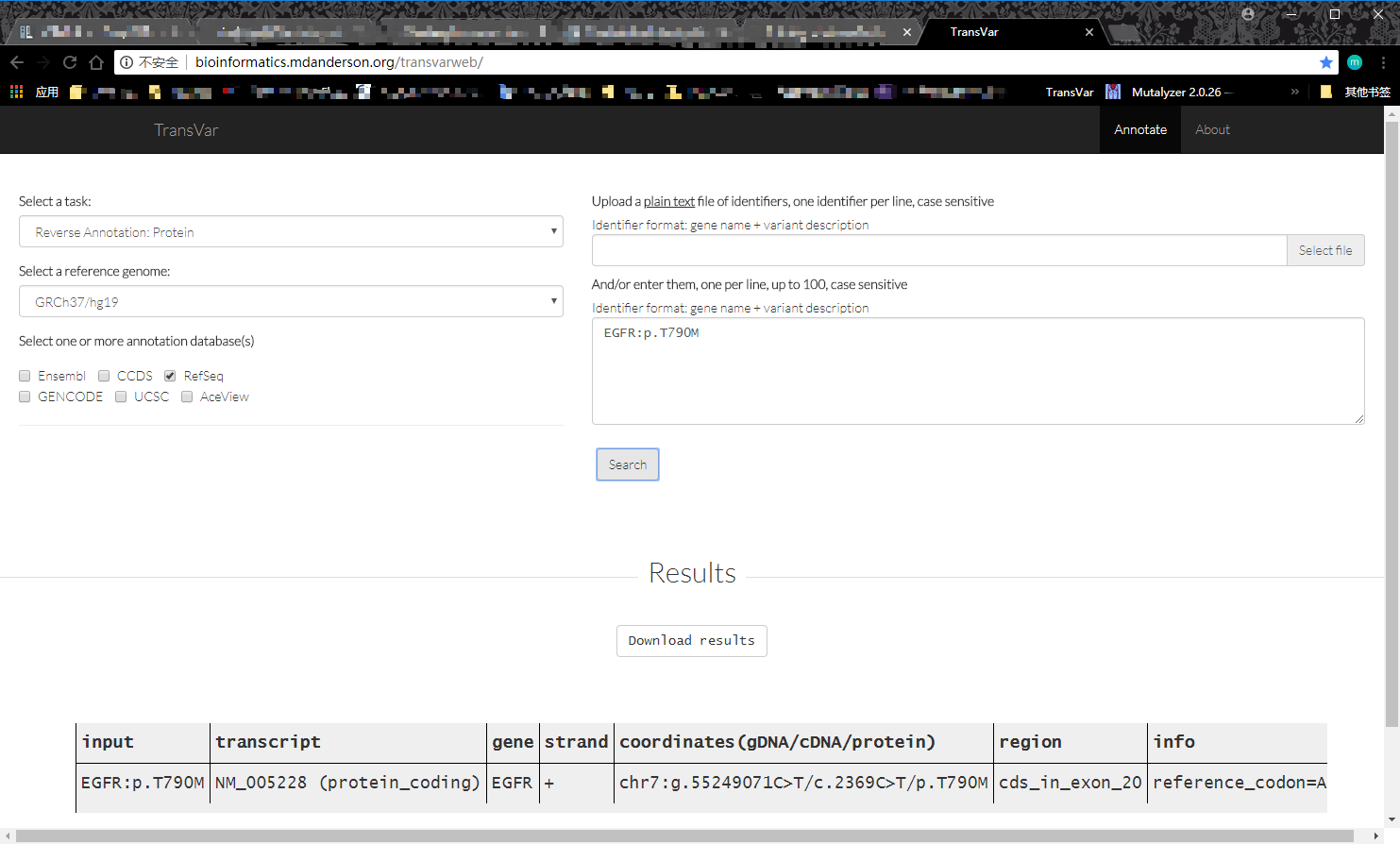
transcvar web 版
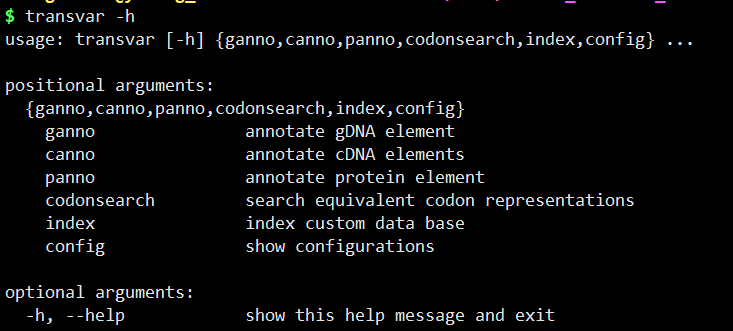
transcvar 本地化
1 | https://github.com/zwdzwd/transvar |
1 | head HGVS_cds |
类似的也可以从 c.HGVS、g.HGVS 开始获得完整注释
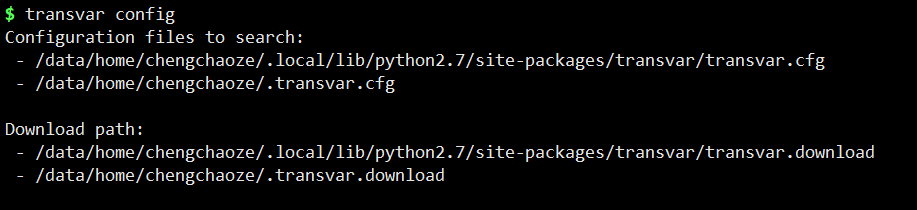
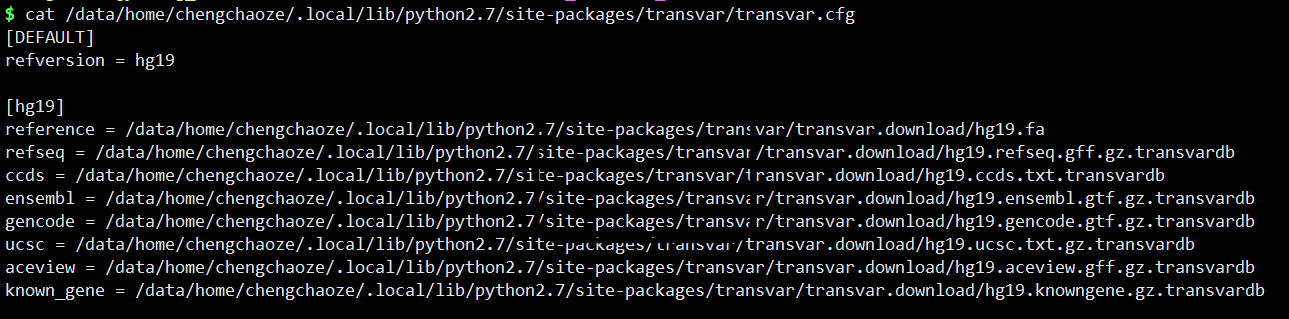
本地化数据库配置
软件提供数据库路径为:http://transvar.info/transvar_user/annotations/,本地数据库配置详细如下:

1 | # https://github.com/zwdzwd/transvar |

1 | head HGVS_cds |
类似的也可以从 c.HGVS、g.HGVS 开始获得完整注释
软件提供数据库路径为:http://transvar.info/transvar_user/annotations/,本地数据库配置详细如下:

