Technical principles
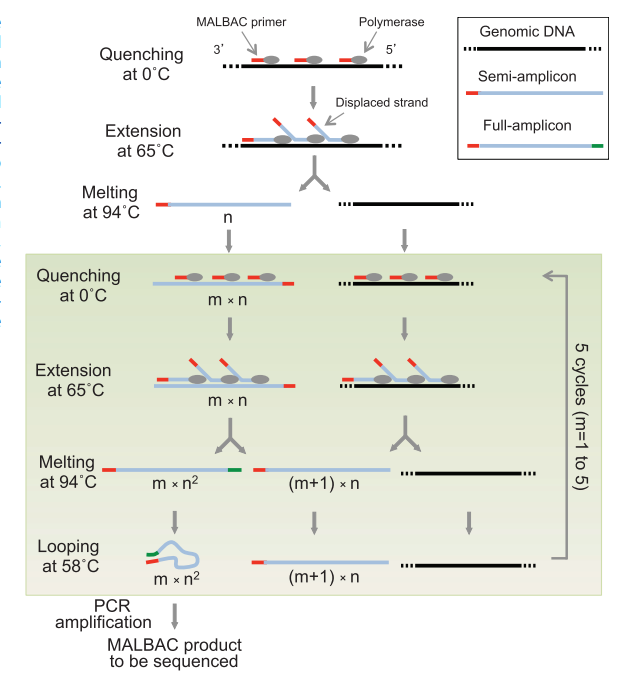
A single cell is picked and lysed. First, genomic DNA of the single cell is melted into single-stranded DNA molecules at 94°C.
Picograms of DNA fragments (~10 to 100 kb)
MALBAC primers then anneal randomly to single-stranded DNA molecules at 0°C and are extended by a polymerase with displacement activity at elevated temperatures, creating semiamplicons.
random primers, each having a common 27-nucleotide sequence and 8 variable nucleotides
In the following five temperature cycles, after the step of looping the full amplicons, singlestranded amplicons and the genomic DNA are used as a template to produce full amplicons and additional semiamplicons, respectively.
For full amplicons, the 3′ end is complementary to the sequence on the 5′ end. The two ends hybridize to form looped DNA, which can efficiently prevent the full amplicon from being used as a template, therefore warranting a close-to-linear amplification.
After the five cycles of linear preamplification, only the full amplicons can be exponentially amplified in the following PCR using the common 27- nucleotide sequence as the primer. PCR reaction will generate microgram level of DNA material for sequencing experiments.
Advantages
- quasi-linear amplification
- accuracy in CNV detection, especially after signal normalization with a reference cell
###Disadvantages
- MALBAC is not free from sequence-dependent bias
- MALBAC is that it has a high false positive rate for SNV detection
- underamplified regions of the genome are sometimes lost during amplification
ref
- 单细胞全基因组扩增技术: MALBAC文章全文翻译
- Zong, Chenghang, et al. “Genome-Wide Detection of Single-Nucleotide and Copy-Number Variations of a Single Human Cell.” Science, vol. 338, no. 6114, 2012, pp. 1622–1626.
- L, Huang, et al. “Single-Cell Whole-Genome Amplification and Sequencing: Methodology and Applications.” Annual Review of Genomics and Human Genetics, vol. 16, no. 1, 2015, pp. 79–102.